Brain Tumor Image Classifier¶
Context¶
In this notebook, we will build an image classifier that can distinguish Pituitary Tumor from "No Tumor" MRI Scan images.
The dataset used in this notebook is available for download from Kaggle.
Although this dataset actually has a total of 3,264 images belonging to 4 classes - Glioma Tumor, Meningioma Tumor, Pituitary Tumor and No Tumor, for this project we have only taken two classes, and we are building a binary classification model to classify between the Pituitary Tumor category vs No Tumor.
For this project, we will only use 1000 of these images (830 training images and 170 Testing images). For the training dataset, we will take 395 MRI scans of No Tumor and 435 MRI scans of Pituitary Tumor. In our problem, we will also be using Data Augmentation to prevent overfitting, and to make our model model more generalised and robust.
We will use this to build an image classification model for this problem statement, and then show how we can improve our performance by simply "importing" a popular pre-trained model architecture and leveraging the idea of Transfer Learning.
Objectives¶
The objectives of this project are to:
- Load and understand the dataset
- Automatically label the images
- Perform Data Augmentation
- Build a classification model for this problem using CNNs
- Improve the model's performance through Transfer Learning
Importing Libraries¶
# Library for creating data paths
import os
# Library for randomly selecting data points
import random
# Library for performing numerical computations
import numpy as np
# Library for creating and showing plots
import matplotlib.pyplot as plt
# Library for reading and showing images
import matplotlib.image as mpimg
# Importing all the required sub-modules from Keras
from keras.models import Sequential, Model
from keras.applications.vgg16 import VGG16
from keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.utils import img_to_array, load_img
from keras.layers import Conv2D, MaxPooling2D, Flatten, Dense, BatchNormalization, Dropout
Mounting the drive to load the dataset
from google.colab import drive
drive.mount('/content/drive')
Mounted at /content/drive
# Unzip the data - We only need to do this once, comment this out if it has already been done
'''
import zipfile
with zipfile.ZipFile('/content/drive/MyDrive/MIT - Data Sciences/Colab Notebooks/Week_Six_-_Deep_Learning/Brain_Tumor_Identification/brain_tumor.zip', 'r') as zip_ref:
zip_ref.extractall('/content/drive/MyDrive/MIT - Data Sciences/Colab Notebooks/Week_Six_-_Deep_Learning/Brain_Tumor_Identification')
'''
"\nimport zipfile\n\nwith zipfile.ZipFile('/content/drive/MyDrive/MIT - Data Sciences/Colab Notebooks/Week_Six_-_Deep_Learning/Brain_Tumor_Identification/brain_tumor.zip', 'r') as zip_ref:\n zip_ref.extractall('/content/drive/MyDrive/MIT - Data Sciences/Colab Notebooks/Week_Six_-_Deep_Learning/Brain_Tumor_Identification') \n"
We have stored the images in a structured folder, and below we create the data paths to load images from those folders. This is required so that we can extract images in an auto-labelled fashion using Keras flow_from_directory.
# Parent directory where images are stored in drive
parent_dir = '/content/drive/MyDrive/MIT - Data Sciences/Colab Notebooks/Week_Six_-_Deep_Learning/Brain_Tumor_Identification/brain_tumor'
# Path to the training and testing datasets within the parent directory
train_dir = os.path.join(parent_dir, 'Training')
validation_dir = os.path.join(parent_dir, 'Testing')
# Directory with our training pictures
train_pituitary_dir = os.path.join(train_dir, 'pituitary_tumor')
train_no_dir = os.path.join(train_dir, 'no_tumor')
# Directory with our testing pictures
validation_pituitary_dir = os.path.join(validation_dir, 'pituitary_tumor')
validation_no_dir = os.path.join(validation_dir, 'no_tumor')
Visualizing a few images¶
Before we move ahead and perform data augmentation, let's randomly check out some of the images and see what they look like:
# Original code that errored out
'''
train_pituitary_file_names = os.listdir(train_pituitary_dir)
train_no_file_names = os.listdir(train_no_dir)
fig = plt.figure(figsize=(16, 8))
fig.set_size_inches(16, 16)
pituitary_img_paths = [os.path.join(train_pituitary_dir, file_name) for file_name in train_pituitary_file_names[:8]]
no_img_paths = [os.path.join(train_no_dir, file_name) for file_name in train_no_file_names[:8]]
for i, img_path in enumerate(pituitary_img_paths + no_img_paths):
ax = plt.subplot(4, 4, i + 1)
ax.axis('Off')
img = mpimg.imread(img_path)
plt.imshow(img)
plt.show()
'''
# Replacement code
train_pituitary_file_names = [f for f in os.listdir(train_pituitary_dir) if not f.startswith('.')] # Filter out files starting with '.'
train_no_file_names = [f for f in os.listdir(train_no_dir) if not f.startswith('.')] # Filter out files starting with '.'
fig = plt.figure(figsize=(16, 8))
fig.set_size_inches(16, 16)
pituitary_img_paths = [os.path.join(train_pituitary_dir, file_name) for file_name in train_pituitary_file_names[:8]]
no_img_paths = [os.path.join(train_no_dir, file_name) for file_name in train_no_file_names[:8]]
for i, img_path in enumerate(pituitary_img_paths + no_img_paths):
ax = plt.subplot(4, 4, i + 1)
ax.axis('Off')
img = mpimg.imread(img_path)
plt.imshow(img)
plt.show()
As we can see, the images are quite different in size from each other.
This represents a problem, as most CNN architectures, including the pre-built model architectures that we will use for Transfer Learning, expect all the images to have the same size.
So we need to crop these images from the center to make sure they all have the same size. We can do this automatically while performing Data Augmentation, as shown below.
Data Augmentation¶
In most real-life case studies, it is generally difficult to collect lots of images and then train CNNs. In that case, one idea we can take advantage of is Data Augmentation. CNNs have the property of translational invariance, i.e., they can recognize an object as an object, even when its appearance varies translationally in some way. Taking this property into consideration, we can augment the images using the following techniques:
1. Horizontal Flip (should be set to True/False)
2. Vertical Flip (should be set to True/False)
3. Height Shift (should be between 0 and 1)
4. Width Shift (should be between 0 and 1)
5. Rotation (should be between 0 and 180)
6. Shear (should be between 0 and 1)
7. Zoom (should be between 0 and 1) etc.
Remember not to use data augmentation in the validation/test data set.
Also, as mentioned above, we need to have images of the same size. So below,we resize the images by using the parameter target_size. Here we are resizing it to 224 x 224, as we will be using the VGG16 model for Transfer Learning, which takes image inputs as 224 x 224.
As this is a binary classification problem, we will need class labels. This is directly handled by the flow_from_directory function. It will take the images from the folder inside our specified directory, and the images from one folder will belong to same class.
As the train directory has 2 folders pituitary_tumor and no_tumor, it will read the directory and each folder will be considered a separate class. We specify class_model = 'binary' as this is a binary classification problem.
As the folders inside the directory will be read in an alphabetical order, the no_tumor folder will be given a label 0, and pituitary_tumor will be label 1.
# All images to be rescaled by 1/255.
train_datagen = ImageDataGenerator(rescale=1. / 255.0,
horizontal_flip = True,
vertical_flip = False,
height_shift_range= 0.1,
width_shift_range=0.1,
rotation_range=20,
shear_range = 0.1,
zoom_range=0.1)
test_datagen = ImageDataGenerator(rescale = 1.0/255.)
# Flowing training images in batches of 20 using train_datagen generator
train_generator = train_datagen.flow_from_directory(train_dir,
batch_size=20,
class_mode='binary',
target_size=(224, 224))
# Flowing testing images in batches of 20 using test_datagen generator
validation_generator = test_datagen.flow_from_directory(validation_dir,
batch_size=20,
class_mode = 'binary',
target_size = (224, 224))
Found 830 images belonging to 2 classes. Found 170 images belonging to 2 classes.
Let's look at some examples of our augmented training data.
This is helpful for understanding the extent to which data is being manipulated prior to training, and can be compared with how the raw data looks prior to data augmentation.
images, labels = next(train_generator)
fig, axes = plt.subplots(4, 4, figsize = (16, 8))
fig.set_size_inches(16, 16)
for (image, label, ax) in zip(images, labels, axes.flatten()):
ax.imshow(image)
if label == 1:
ax.set_title('pituitary tumor')
else:
ax.set_title('no tumor')
ax.axis('off')
CNN Model Building¶
Once the data is augmented and cropped to have the same size, we are now ready to build a first baseline CNN model to classify no_tumor vs pituitary_tumor.
When building our custom model, we have used Batch Normalization and Dropout layers as regularization techniques to prevent overfitting.
cnn_model = Sequential()
cnn_model.add(Conv2D(64, (3,3), activation='relu', input_shape=(224, 224, 3), padding = 'same'))
cnn_model.add(MaxPooling2D(2,2))
cnn_model.add(BatchNormalization())
cnn_model.add(Conv2D(32, (3,3), activation='relu', padding = 'same'))
cnn_model.add(MaxPooling2D(2,2))
cnn_model.add(BatchNormalization())
cnn_model.add(Conv2D(32, (3,3), activation='relu', padding = 'same'))
cnn_model.add(MaxPooling2D(2,2))
cnn_model.add(Conv2D(16, (3,3), activation='relu', padding = 'same'))
cnn_model.add(Flatten())
cnn_model.add(Dense(64, activation='relu'))
cnn_model.add(Dropout(0.25))
cnn_model.add(Dense(32, activation='relu'))
cnn_model.add(Dropout(0.25))
cnn_model.add(Dense(32, activation='relu'))
cnn_model.add(Dense(1, activation='sigmoid'))
cnn_model.compile(loss="binary_crossentropy", optimizer="adam", metrics = ['accuracy'])
cnn_model.summary()
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
conv2d (Conv2D) (None, 224, 224, 64) 1792
max_pooling2d (MaxPooling2 (None, 112, 112, 64) 0
D)
batch_normalization (Batch (None, 112, 112, 64) 256
Normalization)
conv2d_1 (Conv2D) (None, 112, 112, 32) 18464
max_pooling2d_1 (MaxPoolin (None, 56, 56, 32) 0
g2D)
batch_normalization_1 (Bat (None, 56, 56, 32) 128
chNormalization)
conv2d_2 (Conv2D) (None, 56, 56, 32) 9248
max_pooling2d_2 (MaxPoolin (None, 28, 28, 32) 0
g2D)
conv2d_3 (Conv2D) (None, 28, 28, 16) 4624
flatten (Flatten) (None, 12544) 0
dense (Dense) (None, 64) 802880
dropout (Dropout) (None, 64) 0
dense_1 (Dense) (None, 32) 2080
dropout_1 (Dropout) (None, 32) 0
dense_2 (Dense) (None, 32) 1056
dense_3 (Dense) (None, 1) 33
=================================================================
Total params: 840561 (3.21 MB)
Trainable params: 840369 (3.21 MB)
Non-trainable params: 192 (768.00 Byte)
_________________________________________________________________
# Pulling a single large batch of random testing data for testing after each epoch
testX, testY = validation_generator.next()
model_history = cnn_model.fit(train_generator,
validation_data=(testX, testY),
epochs=10)
Epoch 1/10 42/42 [==============================] - 95s 2s/step - loss: 0.5253 - accuracy: 0.7578 - val_loss: 0.6496 - val_accuracy: 0.7000 Epoch 2/10 42/42 [==============================] - 16s 379ms/step - loss: 0.4027 - accuracy: 0.8181 - val_loss: 0.7342 - val_accuracy: 0.6000 Epoch 3/10 42/42 [==============================] - 16s 381ms/step - loss: 0.3711 - accuracy: 0.8386 - val_loss: 1.3765 - val_accuracy: 0.6000 Epoch 4/10 42/42 [==============================] - 16s 381ms/step - loss: 0.3361 - accuracy: 0.8795 - val_loss: 0.6057 - val_accuracy: 0.6000 Epoch 5/10 42/42 [==============================] - 16s 382ms/step - loss: 0.3214 - accuracy: 0.8771 - val_loss: 1.0233 - val_accuracy: 0.6000 Epoch 6/10 42/42 [==============================] - 16s 379ms/step - loss: 0.2688 - accuracy: 0.8952 - val_loss: 1.1162 - val_accuracy: 0.6000 Epoch 7/10 42/42 [==============================] - 16s 383ms/step - loss: 0.1996 - accuracy: 0.9289 - val_loss: 0.3815 - val_accuracy: 0.8000 Epoch 8/10 42/42 [==============================] - 16s 380ms/step - loss: 0.1420 - accuracy: 0.9482 - val_loss: 0.6364 - val_accuracy: 0.7000 Epoch 9/10 42/42 [==============================] - 16s 380ms/step - loss: 0.2579 - accuracy: 0.9072 - val_loss: 0.3263 - val_accuracy: 0.8500 Epoch 10/10 42/42 [==============================] - 16s 382ms/step - loss: 0.2233 - accuracy: 0.9120 - val_loss: 1.5465 - val_accuracy: 0.6500
# Evaluating on the Test dataset
cnn_model.evaluate(validation_generator)
9/9 [==============================] - 36s 4s/step - loss: 1.1214 - accuracy: 0.7353
[1.1214057207107544, 0.7352941036224365]
Findings¶
- Our model had 840,369 trainable parameters.
- After running 10 epochs, we were able to achieve a training accuracy of ~95% but the validation accuracy is comparatively lower than training accuracy.
- Even after using Data Augmentation, Batch Normalization and the Dropout Layers, the model seems to have highly overfit on the training dataset and is performing somewhat poorly.
Model Building using Transfer Learning: VGG 16¶
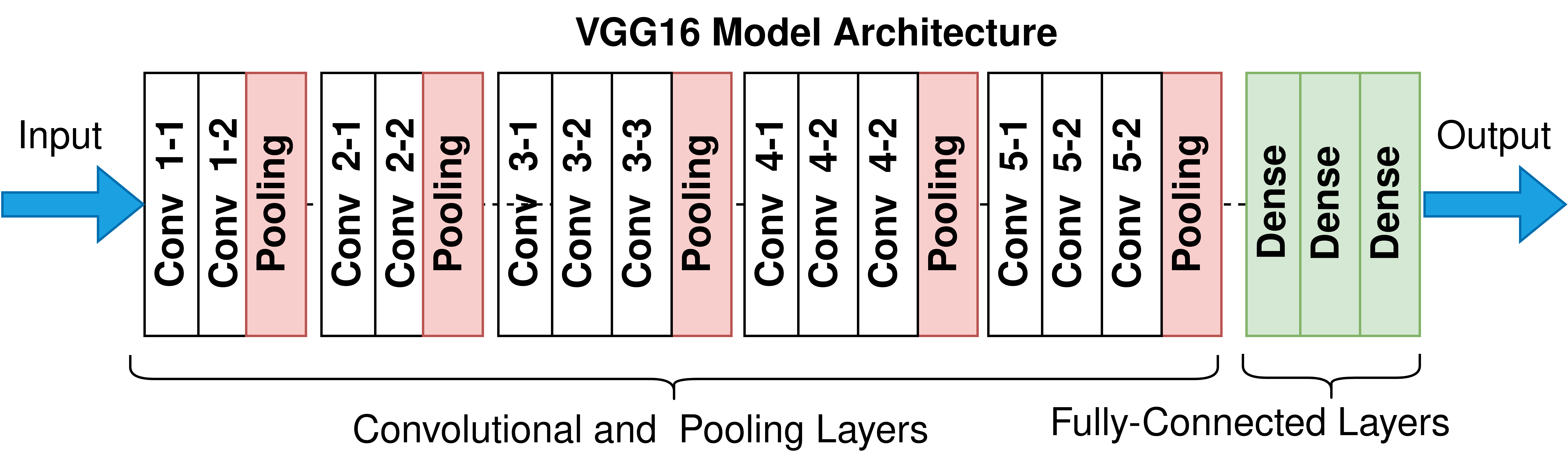
Now, let's try again, but this time, using the idea of Transfer Learning. We will be loading a pre-built architecture - VGG16, which was trained on the ImageNet dataset and finished runner-up in the ImageNet competition in 2014. Below is a schematic of the VGG16 model.
For training VGG16, we will directly use the convolutional and pooling layers and freeze their weights i.e. no training will be done on them. We will remove the already-present fully-connected layers and add our own fully-connected layers for this binary classification task.
# Summary of the whole model
model = VGG16(weights='imagenet')
model.summary()
Downloading data from https://storage.googleapis.com/tensorflow/keras-applications/vgg16/vgg16_weights_tf_dim_ordering_tf_kernels.h5
553467096/553467096 [==============================] - 3s 0us/step
Model: "vgg16"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_1 (InputLayer) [(None, 224, 224, 3)] 0
block1_conv1 (Conv2D) (None, 224, 224, 64) 1792
block1_conv2 (Conv2D) (None, 224, 224, 64) 36928
block1_pool (MaxPooling2D) (None, 112, 112, 64) 0
block2_conv1 (Conv2D) (None, 112, 112, 128) 73856
block2_conv2 (Conv2D) (None, 112, 112, 128) 147584
block2_pool (MaxPooling2D) (None, 56, 56, 128) 0
block3_conv1 (Conv2D) (None, 56, 56, 256) 295168
block3_conv2 (Conv2D) (None, 56, 56, 256) 590080
block3_conv3 (Conv2D) (None, 56, 56, 256) 590080
block3_pool (MaxPooling2D) (None, 28, 28, 256) 0
block4_conv1 (Conv2D) (None, 28, 28, 512) 1180160
block4_conv2 (Conv2D) (None, 28, 28, 512) 2359808
block4_conv3 (Conv2D) (None, 28, 28, 512) 2359808
block4_pool (MaxPooling2D) (None, 14, 14, 512) 0
block5_conv1 (Conv2D) (None, 14, 14, 512) 2359808
block5_conv2 (Conv2D) (None, 14, 14, 512) 2359808
block5_conv3 (Conv2D) (None, 14, 14, 512) 2359808
block5_pool (MaxPooling2D) (None, 7, 7, 512) 0
flatten (Flatten) (None, 25088) 0
fc1 (Dense) (None, 4096) 102764544
fc2 (Dense) (None, 4096) 16781312
predictions (Dense) (None, 1000) 4097000
=================================================================
Total params: 138357544 (527.79 MB)
Trainable params: 138357544 (527.79 MB)
Non-trainable params: 0 (0.00 Byte)
_________________________________________________________________
# Getting only the conv layers for transfer learning.
transfer_layer = model.get_layer('block5_pool')
vgg_model = Model(inputs=model.input, outputs=transfer_layer.output)
vgg_model.summary()
Model: "model"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_1 (InputLayer) [(None, 224, 224, 3)] 0
block1_conv1 (Conv2D) (None, 224, 224, 64) 1792
block1_conv2 (Conv2D) (None, 224, 224, 64) 36928
block1_pool (MaxPooling2D) (None, 112, 112, 64) 0
block2_conv1 (Conv2D) (None, 112, 112, 128) 73856
block2_conv2 (Conv2D) (None, 112, 112, 128) 147584
block2_pool (MaxPooling2D) (None, 56, 56, 128) 0
block3_conv1 (Conv2D) (None, 56, 56, 256) 295168
block3_conv2 (Conv2D) (None, 56, 56, 256) 590080
block3_conv3 (Conv2D) (None, 56, 56, 256) 590080
block3_pool (MaxPooling2D) (None, 28, 28, 256) 0
block4_conv1 (Conv2D) (None, 28, 28, 512) 1180160
block4_conv2 (Conv2D) (None, 28, 28, 512) 2359808
block4_conv3 (Conv2D) (None, 28, 28, 512) 2359808
block4_pool (MaxPooling2D) (None, 14, 14, 512) 0
block5_conv1 (Conv2D) (None, 14, 14, 512) 2359808
block5_conv2 (Conv2D) (None, 14, 14, 512) 2359808
block5_conv3 (Conv2D) (None, 14, 14, 512) 2359808
block5_pool (MaxPooling2D) (None, 7, 7, 512) 0
=================================================================
Total params: 14714688 (56.13 MB)
Trainable params: 14714688 (56.13 MB)
Non-trainable params: 0 (0.00 Byte)
_________________________________________________________________
To remove the fully-connected layers of the imported pre-trained model, while calling it from Keras we can also specify an additonal keyword argument that is include_top.
If we specify include_top = False, then the model will be imported without the fully-connected layers. Here we won't have to do the above steps of getting the last convolutional layer and creating a separate model.
If we are specifying include_top = False, we will also have to specify our input image shape.
Keras has this keyword argument as generally while importing a pre-trained CNN model, we don't require the fully-connected layers and we train our own fully-connected layers for our task.
vgg_model = VGG16(weights='imagenet', include_top = False, input_shape = (224,224,3))
vgg_model.summary()
Downloading data from https://storage.googleapis.com/tensorflow/keras-applications/vgg16/vgg16_weights_tf_dim_ordering_tf_kernels_notop.h5
58889256/58889256 [==============================] - 0s 0us/step
Model: "vgg16"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_2 (InputLayer) [(None, 224, 224, 3)] 0
block1_conv1 (Conv2D) (None, 224, 224, 64) 1792
block1_conv2 (Conv2D) (None, 224, 224, 64) 36928
block1_pool (MaxPooling2D) (None, 112, 112, 64) 0
block2_conv1 (Conv2D) (None, 112, 112, 128) 73856
block2_conv2 (Conv2D) (None, 112, 112, 128) 147584
block2_pool (MaxPooling2D) (None, 56, 56, 128) 0
block3_conv1 (Conv2D) (None, 56, 56, 256) 295168
block3_conv2 (Conv2D) (None, 56, 56, 256) 590080
block3_conv3 (Conv2D) (None, 56, 56, 256) 590080
block3_pool (MaxPooling2D) (None, 28, 28, 256) 0
block4_conv1 (Conv2D) (None, 28, 28, 512) 1180160
block4_conv2 (Conv2D) (None, 28, 28, 512) 2359808
block4_conv3 (Conv2D) (None, 28, 28, 512) 2359808
block4_pool (MaxPooling2D) (None, 14, 14, 512) 0
block5_conv1 (Conv2D) (None, 14, 14, 512) 2359808
block5_conv2 (Conv2D) (None, 14, 14, 512) 2359808
block5_conv3 (Conv2D) (None, 14, 14, 512) 2359808
block5_pool (MaxPooling2D) (None, 7, 7, 512) 0
=================================================================
Total params: 14714688 (56.13 MB)
Trainable params: 14714688 (56.13 MB)
Non-trainable params: 0 (0.00 Byte)
_________________________________________________________________
# Making all the layers of the VGG model non-trainable. i.e. freezing them
for layer in vgg_model.layers:
layer.trainable = False
for layer in vgg_model.layers:
print(layer.name, layer.trainable)
input_2 False block1_conv1 False block1_conv2 False block1_pool False block2_conv1 False block2_conv2 False block2_pool False block3_conv1 False block3_conv2 False block3_conv3 False block3_pool False block4_conv1 False block4_conv2 False block4_conv3 False block4_pool False block5_conv1 False block5_conv2 False block5_conv3 False block5_pool False
new_model = Sequential()
# Adding the convolutional part of the VGG16 model from above
new_model.add(vgg_model)
# Flattening the output of the VGG16 model because it is from a convolutional layer
new_model.add(Flatten())
# Adding a dense output layer
new_model.add(Dense(32, activation='relu'))
new_model.add(Dense(32, activation='relu'))
new_model.add(Dense(1, activation='sigmoid'))
new_model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
new_model.summary()
Model: "sequential_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
vgg16 (Functional) (None, 7, 7, 512) 14714688
flatten_1 (Flatten) (None, 25088) 0
dense_4 (Dense) (None, 32) 802848
dense_5 (Dense) (None, 32) 1056
dense_6 (Dense) (None, 1) 33
=================================================================
Total params: 15518625 (59.20 MB)
Trainable params: 803937 (3.07 MB)
Non-trainable params: 14714688 (56.13 MB)
_________________________________________________________________
## Fitting the VGG model
new_model_history = new_model.fit(train_generator,
validation_data=(testX, testY),
epochs=5)
Epoch 1/5 42/42 [==============================] - 24s 558ms/step - loss: 0.8860 - accuracy: 0.6783 - val_loss: 0.5831 - val_accuracy: 0.8000 Epoch 2/5 42/42 [==============================] - 19s 458ms/step - loss: 0.4458 - accuracy: 0.8831 - val_loss: 0.5458 - val_accuracy: 0.8000 Epoch 3/5 42/42 [==============================] - 19s 454ms/step - loss: 0.3765 - accuracy: 0.9518 - val_loss: 0.5819 - val_accuracy: 0.8000 Epoch 4/5 42/42 [==============================] - 19s 451ms/step - loss: 0.3350 - accuracy: 0.9675 - val_loss: 0.7205 - val_accuracy: 0.8000 Epoch 5/5 42/42 [==============================] - 19s 452ms/step - loss: 0.2925 - accuracy: 0.9699 - val_loss: 0.5999 - val_accuracy: 0.8000
# Evaluating on the Test set
new_model.evaluate(validation_generator)
9/9 [==============================] - 4s 361ms/step - loss: 0.4265 - accuracy: 0.8471
[0.42654949426651, 0.8470588326454163]
# Function to plot loss, val_loss,
def plot_history(history):
N = len(history.history["accuracy"])
plt.figure()
plt.plot(np.arange(0, N), history.history["accuracy"], label="train_accuracy")
plt.plot(np.arange(0, N), history.history["val_accuracy"], label="val_accuracy")
plt.title("Training accuracy Dataset")
plt.xlabel("Epoch #")
plt.ylabel("accuracy")
plt.legend(loc="upper right")
# Plotting the loss vs epoch curve for the basic CNN model without Transfer Learning
plot_history(model_history)
# Plotting the loss vs epoch curve for the Transfer Learning model
plot_history(new_model_history)
Findings¶
- Our model has 803,937 Trainable parameters.
- After running 5 epochs we were able to achieve a good training accuracy and validation accuracy.
Conclusions¶
The difference in both models is evident. Both models had nearly the same number of trainable parameters. However even after training the custom CNN model for 10 epochs, it could not attain accuracies as high as we achieved with Transfer Learning.
The Transfer Learning model has converged faster than the custom CNN model in only 5 epochs.
That's a good level of improvement just by directly using a pre-trained architecture such as VGG16.
This model can, in fact, further be tuned to achieve the accuracies required for practical applicability in the medical domain.
# Convert notebook to html
!jupyter nbconvert --to html "/content/drive/My Drive/Colab Notebooks/Copy of FDS_Project_LearnerNotebook_FullCode.ipynb"